Overview and examples¶
The ChannelPack class is a basic wrapper class for a dict of data and a dict of field names. Those dict attributes, data and names, are a little special – they both require integer keys and the data dict will convert sequence values to Numpy arrays if not arrays already. And the data dict will raise an exception if any resulting array is not 1-dimensional.
The 1-dimensional requirement reflects a view of the ChannelPack object as a holder of flat file data columns.
The integer keys in respective dict are supposed to align to be able to refer to arrays by name.
ChannelPack objects are callable (like functions) and the idea is to get at data by making calls to the object, like pack(ch), where ch is the key for data, either a string name or an integer key.
Make an object¶
ChannelPack takes zero or one dict for data and zero or one dict for names to initialize. data and names can also be assigned after initialization.
Produce some data and make a pack¶
>>> import channelpack as cp
>>> pack = cp.ChannelPack()
>>> pack.data = {0: range(5), 1: ('A', 'B', 'C', 'D', 'E')}
>>> pack.names = {0: 'seq', 1: 'abc'}
>>> pack
ChannelPack(
data={0: array([0, 1, 2, 3, 4]),
1: array(['A', 'B', 'C', 'D', 'E'], dtype='<U1')},
names={0: 'seq',
1: 'abc'})
>>> # make calls to object to get at data
>>> pack(0)
array([0, 1, 2, 3, 4])
>>> pack(0) is pack('seq')
True
The pack is meant to be called to get at data,
(__call__()), but it is not against the
law to operate on the the data and names attributes directly:
>>> pack.data[2] = [letter.lower() for letter in pack('abc')]
>>> pack.names[2] = 'abclower'
>>> pack
ChannelPack(
data={0: array([0, 1, 2, 3, 4]),
1: array(['A', 'B', 'C', 'D', 'E'], dtype='<U1'),
2: array(['a', 'b', 'c', 'd', 'e'], dtype='<U1')},
names={0: 'seq',
1: 'abc',
2: 'abclower'})
Slicing out parts of data¶
Support for slicing and filtering is provided by a Boolean array mask in the pack and the parts or nof arguments in calls. In calls to get at data, the mask is consulted to return parts of the data with corresponding True parts in the mask, depending on arguments. A True entry in the mask represents valid data.
The mask attribute¶
The mask in the pack is set by performing comparisons on arrays, possibly combined with Numpy bitwise operators like & and | (bitwise AND and OR). The goal is to set the mask to a Boolean array of the same size as the data arrays:
>>> pack.mask = (pack('seq') < 2) | (pack('abc') == 'D')
>>> pack('seq', part=0)
array([0, 1])
>>> pack('seq', part=1)
array([3])
>>> pack('abc', nof='filter')
array(['A', 'B', 'D'], dtype='<U1')
>>> pack('abc', nof='nan')
array(['A', 'B', None, 'D', None], dtype=object)
>>> pack('seq', nof='nan')
array([ 0., 1., nan, 3., nan])
- The part argument refer to a contiguous True part of the mask, enumerated from 0. With all elements or only one part True in the mask there is one part == 0. This argument overrides the nof argument.
- With nof=’filter’, a possibly shorter version of data is returned depending on the mask.
- With nof=’nan’, the data length is the same as original array but with corresponding non-true elements in mask replaced with np.nan or None depending on the type.
See also
Start, stop and duration¶
Sometimes it’s easier to think of a part as starting at some event or
condition and stopping at some other. A method
startstop() is supporting something
like a “start and stop trigger”.
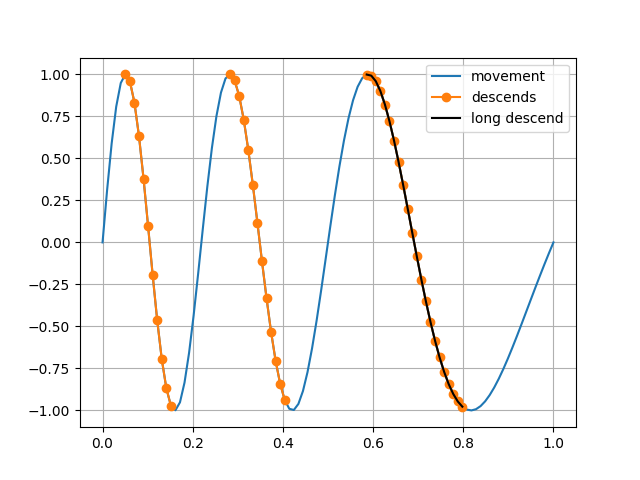
Imagine some alternating movement over time that is slowing down:
>>> import numpy as np
>>> import matplotlib.pyplot as pp
>>> t = np.linspace(0, 1, 100) # (samplerate 100)
>>> f = 5.0
>>> movement = np.sin(2 * np.pi * (f - 2 * t) * t)
>>> pack = cp.ChannelPack({0: t, 1: movement}, {0: 'time', 1: 'movement'})
>>> # Plot the whole movement
>>> _ = pp.plot(pack('time'), pack('movement'), label='movement');
Say that the descending slopes are of particular interest:
>>> startb = pack('movement') > 0.98
>>> stopb = pack('movement') < -0.98
>>> _ = pack.startstop(startb, stopb)
>>> # plot only the descends
>>> _ = pp.plot(pack('time'), pack('movement', nof='nan'),
... label='descends', marker='o')
A method duration() can be used to
make false any true parts that is not long enough. Filter out the
shorter slopes:
>>> _ = pack.duration(0.15, samplerate=100)
>>> # plot only the remaining descend
>>> _ = pp.plot(pack('time'), pack('movement', nof='nan'),
... label='long descend', color='black')
>>> # show it
>>> pp.grid()
>>> _ = pp.legend(loc='upper right'); pp.show()
Factory functions to get a pack¶
A few factory functions are provided to create a pack from data files.
Text¶
Data stored in readable text files in the form of delimited data fields, (csv, txt). Fields might be numbers or text.
If data is numeric only, a lazy variant is available
Spread sheet¶
Code from the library xlrd is used, xls and xlsx types of spread sheets are supported.
